概述
肿瘤细胞游离DNA(cfDNA)存在于癌症患者的血浆中。目前大多数关于循环肿瘤DNA(ctDNA)应用的研究都涉及突变的检测。cfDNA的分析经常在无创检测导致耐药机制的突变以及癌症患者的治疗和疾病监测的背景下讨论。事实上,这一领域已经取得了实质性的进展,开发出了能够达到高灵敏度并能探询大量基因的方法。然而,有趣的是,cfDNA还可以用于分析DNA的不同特征,如甲基化状态、大小片段模式、转录组学和病毒载量,这为癌症患者液体活检样本的分析开辟了新的途径。本文就实体恶性肿瘤患者cfDNA突变检测的新观点和挑战作一综述。
肺癌病友交流群

肿瘤细胞游离DNA(即无细胞DNA,英文缩写cfDNA)是指源自体液中任何细胞类型的细胞外DNA分子(双链DNA和线粒体DNA)。cfDNA已于1948年在患病和健康个体的血液中被检测到。cfDNA分析目前已用于产前诊断,其临床应用也在癌症、器官移植、自身免疫性疾病、创伤、心肌梗死和败血症等领域进行了评估。但是,我们对cfDNA的结构和起源,细胞释放机制和清除的了解仍是初步的。尽管大多数cfDNA分子起源于造血系统,但人们对确定健康和病理条件下不同器官对cfDNA总量的相对贡献有着极大的兴趣。不仅有多种释放机制,包括凋亡,衰老,肥大症,NETNET,吞噬作用和坏死,还包括主动分泌-包括与细胞外囊泡的结合或由其他机制(如红细胞释放成熟核,线粒体DNA分泌或重要的NETosis)诱导。另一方面,不同的参数控制着cfDNA分子的降解和消除:循环中的酶促裂解,通过肝脏消除核小体复合物,并在较小程度上通过肾脏除去DNA片段。cfDNA生物学的这些基本方面的描述超出了本文讨论的范围,故不做过多阐述。
cfDNA的肿瘤来源部分,通常称为循环肿瘤DNA(ctDNA),由于其作为癌症患者微创肿瘤生物标志物的巨大潜力,在过去十年中受到了广泛关注。至于cfDNA,肿瘤生物学与ctDNA释放之间的相关性仍未得到很好的理解,并且可能不仅仅依赖于垂死细胞的数量。不仅肿瘤的体积和代谢,而且其扩散速率也与血浆中ctDNA的含量呈正相关。
关于ctDNA在肿瘤学已发表研究阐明的潜在用途中,绝大多数提及的是在癌症患者血浆或血清中检测到的特定突变。简言之,ctDNA突变检测有可能用于早期癌症检测,判断组织起源、预后,监测疗效和评估治疗的潜在耐药性,或检测微小残留疾病。但是,表观遗传学改变在癌症发展中比体细胞突变更为常见。尽管ctDNA的突变分析显示了许多临床应用,但对cfDNA的评估超出了点突变的检测范围,包括对染色体重排,拷贝数畸变,甲基化,片段化和基因表达的研究,因此也越来越引起人们的关注(图1)。
图1:ctDNA的不同特征及其潜在的临床意义。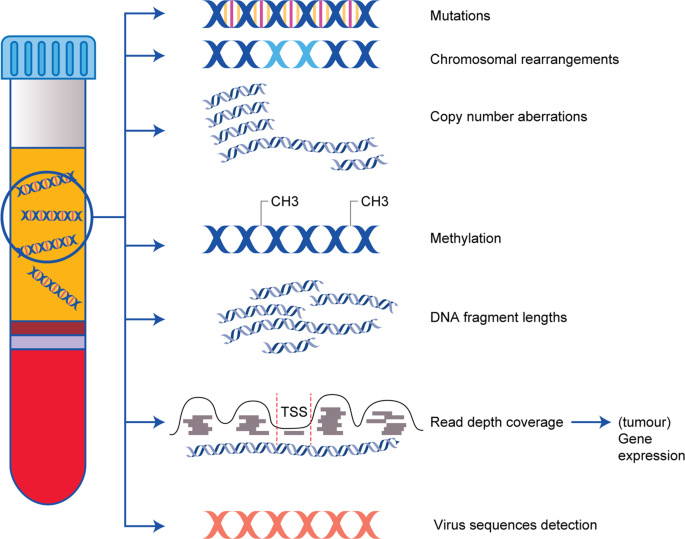
该图总结了可从cfDNA不同特征的研究中获得的与肿瘤相关的临床信息。ctDNA上可检测到的体细胞基因组畸变包括突变,染色体重排和拷贝数畸变。ctDNA的其他特征是特定的表观遗传畸变,例如甲基化模式或不同的DNA片段长度。通过读取核小体间的深度覆盖范围,也可以从ctDNA分析中获得有关肿瘤特异性转录的信息。在病毒引起的肿瘤(例如EBV相关的鼻咽癌或HPV相关的头颈部肿瘤)中,病毒序列的定量评估具有诊断意义。
显然,一些肿瘤类型和身体部位向血液中释放的ctDNA量较低。在这里,用于分子分析的非血液来源的ctDNA已经变得很有价值。很明显,在原发性脑肿瘤,如胶质瘤、中枢神经系统淋巴瘤和一些儿科实体瘤中,脑脊液(CSF)显示出比外周血更高的敏感性。同样,对于一些上呼吸道系统的肿瘤,唾液、痰或胸腔积液也可能是血液的良好替代物,最近的一些研究结果发现,尿液、粪便和精液等其他体液或样本,也适用于不同的液体活检方法。
结论与观点
越来越多的数据表明,除了从癌症患者血浆中获得的cfDNA突变外,还可能获得更多的信息,例如从片段模式或甲基化状态的分析中获得的信息,这些信息对于基因表达的调控尤其有用。人类恶性肿瘤细胞表现出DNA甲基化模式的普遍变化,从而导致基因表达的紊乱或基因组的不稳定性。从癌症治疗的潜在临床前景来看,从早期癌症检测到估计预后和监测治疗反应,破译这些异常的表观遗传修饰是至关重要的。对cfDNA的研究也显示了早期检测与病毒相关癌症的临床潜力,与实体肿瘤中复杂的体细胞突变谱相比,利用不同致病性病毒dna的复杂性会较低些。然而,剩下的挑战将是区分暂时性病毒感染和导致癌症的持续性感染。病毒ctDNA序列的检测还可以为血浆cfDNA的生物学和动力学提供重要的基础信息。对接受鼻咽切除术的鼻咽癌患者血浆中EBV负荷的连续监测显示,血浆中EBV cfDNA的清除率符合一级衰变动力学模型,中位半衰期仅为139 min。数据显示,EBV-DNA的清除非常迅速,因此与基线测量相比,手术后抽血可能是疾病复发的更好预测指标。将cfDNA分析引入癌症诊断的一个重要前提是将现有cfDNA技术的分析前和分析变量标准化。为此,已经建立了包括来自学术界、商界的合作伙伴在内的国际财团组织,例如CANCER-ID或ELBS,并且已经进行了环实验(在多个地点并行使用相同的样品或方法)。此外,越来越多的数据表明,其他基于唾液、脑脊液或尿液的非血源性液体活检方法可用于未来的临床试验。最后,值得一提的是,其他液体活检分析物,如循环肿瘤细胞、循环microRNAs、肿瘤培养血小板或肿瘤相关蛋白,可能提供肿瘤演变和癌症患者治疗反应的补充信息,开发一个复杂的多分析物生物标志物小组,这将需要复杂的生物信息学工具,如机器学习算法,将大大有助于癌症患者个体的无创治疗。
综上所述,10年前提出的液体活检的概念为癌症诊断开辟了新的途径,现在需要进行具有既定疗效指标的介入性临床试验,以进一步证明ctDNA和其他生物标记物的临床效用。
参考文献
1. 1.Mandel, P. & Metais, P. Les acides nucléiques du plasma sanguin chez l’homme. Comptes rendus des. seances de. la Soc. de. biologie et. de. ses. filiales 142, 241–243 (1948)2. 2.Lo, Y. M., Chan, K. C., Sun, H., Chen, E. Z., Jiang, P., Lun, F. M. et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2, 61ra91 (2010).
3. 3.Knight, S. R., Thorne, A. & Lo Faro, M. L. Donor-specific cell-free DNA as a biomarker in solid organ transplantation. A systematic review. Transplantation 103, 273–283 (2019).
4. 4.Duvvuri, B. & Lood, C. Cell-Free DNA as a biomarker in autoimmune rheumatic diseases. Front. Immunol. 10, 502 (2019).
5. 5.Lehmann-Werman, R., Magenheim, J., Moss, J., Neiman, D., Abraham, O., Piyanzin, S. et al. Monitoring liver damage using hepatocyte-specific methylation markers in cell-free circulating DNA. JCI Insight 3, e120687 (2018).
6. 6.Zemmour, H., Planer, D., Magenheim, J., Moss, J., Neiman, D., Gilon, D. et al. Non-invasive detection of human cardiomyocyte death using methylation patterns of circulating DNA. Nat. Commun. 9, 1443 (2018).
7. 7.Wan, J. C. M., Massie, C., Garcia-Corbacho, J., Mouliere, F., Brenton, J. D., Caldas, C. et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 (2017).
8. 8.van der Pol, Y. & Mouliere, F. Toward the early detection of cancer by decoding the epigenetic and environmental fingerprints of cell-free DNA. Cancer Cell. 36, 350–368 (2019).
9. 9.Kustanovich, A., Schwartz, R., Peretz, T. & Grinshpun, A. Life and death of circulating cell-free DNA. Cancer Biol. Ther. 20, 1057–1067 (2019).
10. 10.Abbosh, C., Birkbak, N. J., Wilson, G. A., Jamal-Hanjani, M., Constantin, T., Salari, R. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017).
CAS PubMed PubMed Central Article Google Scholar
11. 11.Mair, R., Mouliere, F., Smith, C. G., Chandrananda, D., Gale, D., Marass, F. et al. Measurement of plasma cell-free mitochondrial tumor DNA improves detection of glioblastoma in patient-derived orthotopic xenograft models. Cancer Res. 79, 220–230 (2019).
CAS PubMed Article PubMed Central Google Scholar
12. 12.Wong, S. Q., Raleigh, J. M., Callahan, J., Vergara, I. A., Ftouni, S., Hatzimihalis, A. et al. Circulating tumor DNA analysis and functional imaging provide complementary approaches for comprehensive disease monitoring in metastatic melanoma. JCO Precis. Oncol. 1, 1–14 (2017).
CAS Google Scholar
13. 13.Lazaro-Ibanez, E., Lasser, C., Shelke, G. V., Crescitelli, R., Jang, S. C., Cvjetkovic, A. et al. DNA analysis of low- and high-density fractions defines heterogeneous subpopulations of small extracellular vesicles based on their DNA cargo and topology. J. Extracell. Vesicles 8, 1656993 (2019).
CAS PubMed PubMed Central Article Google Scholar
14. 14.Vagner, T., Spinelli, C., Minciacchi, V. R., Balaj, L., Zandian, M., Conley, A. et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J. Extracell. Vesicles 7, 1505403 (2018).
CAS PubMed PubMed Central Article Google Scholar
15. 15.Heitzer, E., Haque, I. S., Roberts, C. E. S. & Speicher, M. R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 20, 71–88 (2019).
CAS PubMed Article PubMed Central Google Scholar
16. 16.Feinberg, A. P., Koldobskiy, M. A. & Gondor, A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet. 17, 284–299 (2016).
CAS PubMed PubMed Central Article Google Scholar
17. 17.Boire, A., Brandsma, D., Brastianos, P. K., Le Rhun, E., Ahluwalia, M., Junck, L. et al. Liquid biopsy in central nervous system metastases: a RANO review and proposals for clinical applications. Neuro Oncol. 21, 571–584 (2019).
CAS PubMed PubMed Central Article Google Scholar
18. 18.Abbou, S. D., Shulman, D. S., DuBois, S. G. & Crompton, B. D. Assessment of circulating tumor DNA in pediatric solid tumors: the promise of liquid biopsies. Pediatr. Blood Cancer 66, e27595 (2019).
PubMed PubMed Central Article Google Scholar
19. 19.Cristaldi, M., Mauceri, R., Di Fede, O., Giuliana, G., Campisi, G. & Panzarella, V. Salivary biomarkers for oral squamous cell carcinoma diagnosis and follow-up: current status and perspectives. Front. Physiol. 10, 1476 (2019).
PubMed PubMed Central Article Google Scholar
20. 20.Ribeiro, I. P., de Melo, J. B. & Carreira, I. M. Head and neck cancer: searching for genomic and epigenetic biomarkers in body fluids—the state of art. Mol. Cytogenet. 12, 33 (2019).
PubMed PubMed Central Article CAS Google Scholar
21. 21.Ponti, G., Manfredini, M. & Tomasi, A. Non-blood sources of cell-free DNA for cancer molecular profiling in clinical pathology and oncology. Crit. Rev. Oncol. Hematol. 141, 36–42 (2019).
PubMed Article PubMed Central Google Scholar
22. 22.Elazezy, M. & Joosse, S. A. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput. Struct. Biotechnol. J. 16, 370–378 (2018).
CAS PubMed PubMed Central Article Google Scholar
23. 23.Kwapisz, D. The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Ann. Transl. Med. 5, 46 (2017).
PubMed PubMed Central Article Google Scholar
24. 24.Haselmann, V., Gebhardt, C., Brechtel, I., Duda, A., Czerwinski, C., Sucker, A. et al. Liquid profiling of circulating tumor DNA in plasma of melanoma patients for companion diagnostics and monitoring of BRAF inhibitor therapy. Clin. Chem. 64, 830–42. (2018).
CAS PubMed Article PubMed Central Google Scholar
25. 25.Oshiro, C., Kagara, N., Naoi, Y., Shimoda, M., Shimomura, A., Maruyama, N. et al. PIK3CA mutations in serum DNA are predictive of recurrence in primary breast cancer patients. Breast Cancer Res. Treat. 150, 299–307 (2015).
PubMed Article PubMed Central Google Scholar
26. 26.Kuo, Y. B., Chen, J. S., Fan, C. W., Li, Y. S. & Chan, E. C. Comparison of KRAS mutation analysis of primary tumors and matched circulating cell-free DNA in plasmas of patients with colorectal cancer. Clin. Chim. Acta 433, 284–289 (2014).
CAS PubMed Article PubMed Central Google Scholar
27. 27.Ou, C.-Y., Vu, T., Grunwald, J. T., Toledano, M., Zimak, J., Toosky, M. et al. An ultrasensitive test for profiling circulating tumor DNA using integrated comprehensive droplet digital detection. Lab Chip 19, 993–1005 (2019).
CAS PubMed Article PubMed Central Google Scholar
28. 28.Pantel, K. & Alix-Panabieres, C. Liquid biopsy and minimal residual disease—latest advances and implications for cure. Nat. Rev. Clin. Oncol. 16, 409–24. (2019).
CAS PubMed Article PubMed Central Google Scholar
29. 29.Tan, L., Sandhu, S., Lee, R. J., Li, J., Callahan, J., Ftouni, S. et al. Prediction and monitoring of relapse in stage III melanoma using circulating tumor DNA. Ann. Oncol. 30, 804–14. (2019).
CAS PubMed PubMed Central Article Google Scholar
30. 30.Lee, J. H., Saw, R. P., Thompson, J. F., Lo, S., Spillane, A. J., Shannon, K. F. et al. Pre-operative ctDNA predicts survival in high-risk stage III cutaneous melanoma patients. Ann. Oncol. 30, 815–22. (2019).
CAS PubMed PubMed Central Article Google Scholar
31. 31.Rothwell, D. G., Ayub, M., Cook, N., Thistlethwaite, F., Carter, L., Dean, E. et al. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat. Med. 25, 738–43. (2019).
CAS PubMed Article PubMed Central Google Scholar
32. 32.Gray, E. S., Witkowski, T., Pereira, M., Calapre, L., Herron, K., Irwin, D. et al. Genomic analysis of circulating tumor DNA using a melanoma-specific UltraSEEK oncogene panel. J. Mol. Diagn. 21, 418–26. (2019).
CAS PubMed Article PubMed Central Google Scholar
33. 33.Gremel, G., Lee, R. J., Girotti, M. R., Mandal, A. K., Valpione, S., Garner, G. et al. Distinct subclonal tumour responses to therapy revealed by circulating cell-free DNA. Ann. Oncol. 27, 1959–1965 (2016).
CAS PubMed PubMed Central Article Google Scholar
34. 34.Parikh, A. R., Leshchiner, I., Elagina, L., Goyal, L., Levovitz, C., Siravegna, G. et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 25, 1415–21. (2019).
CAS PubMed PubMed Central Article Google Scholar
35. 35.Forschner, A., Battke, F., Hadaschik, D., Schulze, M., Weissgraeber, S., Han, C. T. et al. Tumor mutation burden and circulating tumor DNA in combined CTLA-4 and PD-1 antibody therapy in metastatic melanoma—results of a prospective biomarker study. J. Immunother. Cancer 7, 180 (2019).
PubMed PubMed Central Article Google Scholar
36. 36.Chae, Y. K., Davis, A. A., Agte, S., Pan, A., Simon, N. I., Iams, W. T. et al. Clinical implications of circulating tumor DNA tumor mutational burden (ctDNA TMB) in non-small cell lung cancer. Oncologist 24, 820–828 (2019).
CAS PubMed PubMed Central Article Google Scholar
37. 37.Hofman, P., Heeke, S., Alix-Panabieres, C. & Pantel, K. Liquid biopsy in the era of immuno-oncology: is it ready for prime-time use for cancer patients? Ann. Oncol. 30, 1448–59. (2019).
CAS PubMed Article PubMed Central Google Scholar
38. 38.Cabel, L., Proudhon, C., Romano, E., Girard, N., Lantz, O., Stern, M. H. et al. Clinical potential of circulating tumour DNA in patients receiving anticancer immunotherapy. Nat. Rev. Clin. Oncol. 15, 639–50. (2018).
CAS PubMed Article PubMed Central Google Scholar
39. 39.Abbosh, C., Birkbak, N. J. & Swanton, C. Early stage NSCLC—challenges to implementing ctDNA-based screening and MRD detection. Nat. Rev. Clin. Oncol. 15, 577–86. (2018).
CAS PubMed Article PubMed Central Google Scholar
40. 40.Bettegowda, C., Sausen, M., Leary, R. J., Kinde, I., Wang, Y., Agrawal, N. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra24 (2014).
PubMed PubMed Central Article CAS Google Scholar
41. 41.Thierry, A. R., Mouliere, F., El Messaoudi, S., Mollevi, C., Lopez-Crapez, E., Rolet, F. et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat. Med. 20, 430–435 (2014).
CAS PubMed Article PubMed Central Google Scholar
42. 42.Parkinson, C. A., Gale, D., Piskorz, A. M., Biggs, H., Hodgkin, C., Addley, H. et al. Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: a retrospective study. PLoS Med. 13, e1002198 (2016).
PubMed PubMed Central Article CAS Google Scholar
43. 43.Otandault, A., Abraham, J. D., Al Amir Dache, Z., Khalyfa, A., Jariel-Encontre, I., Forne, T. et al. Hypoxia differently modulates the release of mitochondrial and nuclear DNA. Br. J. Cancer. 122, 715–725 (2020).
CAS PubMed PubMed Central Article Google Scholar
44. 44.Labgaa, I., Villacorta-Martin, C., D’Avola, D., Craig, A. J., von Felden, J., Martins-Filho, S. N. et al. A pilot study of ultra-deep targeted sequencing of plasma DNA identifies driver mutations in hepatocellular carcinoma. Oncogene 37, 3740–3752 (2018).
CAS PubMed PubMed Central Article Google Scholar
45. 45.Reinert, T., Henriksen, T. V., Christensen, E., Sharma, S., Salari, R., Sethi, H. et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol 5, 1124–1131 (2019).
PubMed PubMed Central Article Google Scholar
46. 46.Christensen, E., Birkenkamp-Demtroder, K., Sethi, H., Shchegrova, S., Salari, R., Nordentoft, I. et al. Early detection of metastatic relapse and monitoring of therapeutic efficacy by ultra-deep sequencing of plasma cell-free DNA in patients with urothelial bladder carcinoma. J. Clin. Oncol. 37, 1547–1557 (2019).
CAS PubMed Article PubMed Central Google Scholar
47. 47.Razavi, P., Li, B. T., Brown, D. N., Jung, B., Hubbell, E., Shen, R. et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 25, 1928–1937 (2019).
CAS PubMed PubMed Central Article Google Scholar
48. 48.Sanchez, C., Snyder, M. W., Tanos, R., Shendure, J. & Thierry, A. R. New insights into structural features and optimal detection of circulating tumor DNA determined by single-strand DNA analysis. npj Genom. Med. 3, 31 (2018).
PubMed PubMed Central Article CAS Google Scholar
49. 49.Mansukhani, S., Barber, L. J., Kleftogiannis, D., Moorcraft, S. Y., Davidson, M., Woolston, A. et al. Ultra-sensitive mutation detection and genome-wide DNA copy number reconstruction by error-corrected circulating tumor DNA sequencing. Clin. Chem. 64, 1626–1635 (2018).
CAS PubMed PubMed Central Article Google Scholar
50. 50.Odegaard, J. I., Vincent, J. J., Mortimer, S., Vowles, J. V., Ulrich, B. C., Banks, K. C. et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin. Cancer Res. 24, 3539–3549 (2018).
CAS PubMed Article PubMed Central Google Scholar
51. 51.Merker, J. D., Oxnard, G. R., Compton, C., Diehn, M., Hurley, P., Lazar, A. J. et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 36, 1631–1641 (2018).
CAS PubMed Article PubMed Central Google Scholar
52. 52.Strickler, J. H., Loree, J. M., Ahronian, L. G., Parikh, A. R., Niedzwiecki, D., Pereira, A. A. L. et al. Genomic landscape of cell-free DNA in patients with colorectal cancer. Cancer Discov. 8, 164–173 (2018).
CAS PubMed Article PubMed Central Google Scholar
53. 53.Stetson, D., Ahmed, A., Xu, X., Nuttall, B. R. B., Lubinski, T. J., Johnson, J. H. et al. Orthogonal comparison of four plasma NGS tests with tumor suggests technical factors are a major source of assay discordance. JCO Precis. Oncol. 3, 1–9 (2019).
Google Scholar
54. 54.Paweletz, C. P., Lau, C. J. & Oxnard, G. R. Does testing error underlie liquid biopsy discordance? JCO Precis. Oncol. 3, 1–3 (2019).
Google Scholar
55. 55.Keller, L. & Pantel, K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat. Rev. Cancer 19, 553–567 (2019).
CAS PubMed Article PubMed Central Google Scholar
56. 56.Chan, K. C., Jiang, P., Zheng, Y. W., Liao, G. J., Sun, H., Wong, J. et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin. Chem. 59, 211–224 (2013).
CAS PubMed Article PubMed Central Google Scholar
57. 57.Murtaza, M., Dawson, S. J., Pogrebniak, K., Rueda, O. M., Provenzano, E., Grant, J. et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat. Commun. 6, 8760 (2015).
PubMed PubMed Central Article CAS Google Scholar
58. 58.Zhang, Y., Chang, L., Yang, Y., Fang, W., Guan, Y., Wu, A. et al. Intratumor heterogeneity comparison among different subtypes of non-small-cell lung cancer through multi-region tissue and matched ctDNA sequencing. Mol. Cancer 18, 7 (2019).
PubMed PubMed Central Article Google Scholar
59. 59.Zhao, M., Wang, Q., Wang, Q., Jia, P. & Zhao, Z. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: features and perspectives. BMC Bioinforma. 14, S1 (2013).
Article Google Scholar
60. 60.Redon, R., Ishikawa, S., Fitch, K. R., Feuk, L., Perry, G. H., Andrews, T. D. et al. Global variation in copy number in the human genome. Nature 444, 444–454 (2006).
CAS PubMed PubMed Central Article Google Scholar
61. 61.Leary, R. J., Sausen, M., Kinde, I., Papadopoulos, N., Carpten, J. D., Craig, D. et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci. Transl. Med. 4, 162ra54 (2012).
Article CAS Google Scholar
62. 62.Heitzer, E., Ulz, P., Belic, J., Gutschi, S., Quehenberger, F., Fischereder, K. et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 5, 30 (2013).
CAS PubMed PubMed Central Article Google Scholar
63. 63.Adalsteinsson, V. A., Ha, G., Freeman, S. S., Choudhury, A. D., Stover, D. G., Parsons, H. A. et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 8, 1324 (2017).
PubMed PubMed Central Article CAS Google Scholar
64. 64.Kirkizlar, E., Zimmermann, B., Constantin, T., Swenerton, R., Hoang, B., Wayham, N. et al. Detection of clonal and subclonal copy-number variants in cell-free DNA from patients with breast cancer using a massively multiplexed PCR methodology. Transl. Oncol. 8, 407–416 (2015).
PubMed PubMed Central Article Google Scholar
65. 65.Douville, C., Springer, S., Kinde, I., Cohen, J. D., Hruban, R. H., Lennon, A. M. et al. Detection of aneuploidy in patients with cancer through amplification of long interspersed nucleotide elements (LINEs). Proc. Natl Acad. Sci. USA 115, 1871–1876 (2018).
CAS PubMed Article PubMed Central Google Scholar
66. 66.Jiang, P., Chan, C. W. M., Chan, K. C. A., Cheng, S. H., Wong, J., Wong, V. W.-S. et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc. Natl Acad. Sci. USA 112, E1317–E1325 (2015).
CAS PubMed Article PubMed Central Google Scholar
67. 67.Chen, X., Chang, C. W., Spoerke, J. M., Yoh, K. E., Kapoor, V., Baudo, C. et al. Low-pass whole-genome sequencing of circulating cell-free DNA demonstrates dynamic changes in genomic copy number in a squamous lung cancer clinical cohort. Clin. Cancer Res. 25, 2254–2263 (2019).
CAS PubMed Article PubMed Central Google Scholar
68. 68.Belic, J., Graf, R., Bauernhofer, T., Cherkas, Y., Ulz, P., Waldispuehl-Geigl, J. et al. Genomic alterations in plasma DNA from patients with metastasized prostate cancer receiving abiraterone or enzalutamide. Int. J. Cancer 143, 1236–1248 (2018).
CAS PubMed PubMed Central Article Google Scholar
69. 69.Pailler, E., Oulhen, M., Borget, I., Remon, J., Ross, K., Auger, N. et al. Circulating tumor cells with aberrant ALK copy number predict progression-free survival during crizotinib treatment in ALK-rearranged non-small cell lung cancer patients. Cancer Res. 77, 2222–2230 (2017).
CAS PubMed Article PubMed Central Google Scholar
70. 70.Pailler, E., Auger, N., Lindsay, C. R., Vielh, P., Islas-Morris-Hernandez, A., Borget, I. et al. High level of chromosomal instability in circulating tumor cells of ROS1-rearranged non-small-cell lung cancer. Ann. Oncol. 26, 1408–1415 (2015).
CAS PubMed PubMed Central Article Google Scholar
71. 71.Dagogo-Jack, I., Rooney, M., Nagy, R. J., Lin, J. J., Chin, E., Ferris, L. A. et al. Molecular analysis of plasma from patients with ROS1-positive NSCLC. J. Thorac. Oncol. 14, 816–824 (2019).
CAS PubMed PubMed Central Article Google Scholar
72. 72.Dagogo-Jack, I., Brannon, A. R., Ferris, L. A., Campbell, C. D., Lin, J. J., Schultz, K. R. et al. Tracking the evolution of resistance to ALK tyrosine kinase inhibitors through longitudinal analysis of circulating tumor DNA. JCO Precis. Oncol. (2018). https://doi.org/10.1200/PO.17.00160.
73. 73.Jahr, S., Hentze, H., Englisch, S., Hardt, D., Fackelmayer, F. O., Hesch, R. D. et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 61, 1659–1665 (2001).
CAS PubMed PubMed Central Google Scholar
74. 74.Giacona, M. B., Ruben, G. C., Iczkowski, K. A., Roos, T. B., Porter, D. M. & Sorenson, G. D. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas 17, 89–97 (1998).
CAS PubMed Article PubMed Central Google Scholar
75. 75.Nagata, S. Apoptotic DNA fragmentation. Exp. Cell Res. 256, 12–18 (2000).
CAS PubMed Article PubMed Central Google Scholar
76. 76.Wang, B. G., Huang, H. Y., Chen, Y. C., Bristow, R. E., Kassauei, K., Cheng, C. C. et al. Increased plasma DNA integrity in cancer patients. Cancer Res. 63, 3966–3968 (2003).
CAS PubMed PubMed Central Google Scholar
77. 77.Underhill, H. R., Kitzman, J. O., Hellwig, S., Welker, N. C., Daza, R., Baker, D. N. et al. Fragment length of circulating tumor DNA. PLoS Genet. 12, e1006162 (2016).
PubMed PubMed Central Article CAS Google Scholar
78. 78.Mouliere, F., Chandrananda, D., Piskorz, A. M., Moore, E. K., Morris, J., Ahlborn, L. B. et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 10, eaat4921 (2018).
PubMed PubMed Central Article CAS Google Scholar
79. 79.Mouliere, F., Robert, B., Arnau Peyrotte, E., Del Rio, M., Ychou, M., Molina, F. et al. High fragmentation characterizes tumour-derived circulating DNA. PloS ONE 6, e23418 (2011).
CAS PubMed PubMed Central Article Google Scholar
80. 80.Cristiano, S., Leal, A., Phallen, J., Fiksel, J., Adleff, V., Bruhm, D. C. et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570, 385–389 (2019).
CAS PubMed PubMed Central Article Google Scholar
81. 81.Bronkhorst, A. J., Wentzel, J. F., Aucamp, J., van Dyk, E., du Plessis, L. & Pretorius, P. J. Characterization of the cell-free DNA released by cultured cancer cells. Biochim. Biophys. Acta 1863, 157–165 (2016).
CAS PubMed Article PubMed Central Google Scholar
82. 82.Norris, A. L., Workman, R. E., Fan, Y., Eshleman, J. R. & Timp, W. Nanopore sequencing detects structural variants in cancer. Cancer Biol. Ther. 17, 246–253 (2016).
CAS PubMed PubMed Central Article Google Scholar
83. 83.Mouliere, F., Mair, R., Chandrananda, D., Marass, F., Smith, C. G., Su, J. et al. Detection of cell-free DNA fragmentation and copy number alterations in cerebrospinal fluid from glioma patients. EMBO Mol. Med. 10, e9323 (2018).
PubMed PubMed Central Article CAS Google Scholar
84. 84.Markus, H., Zhao, J., Contente-Cuomo, T., Raupach, E., Odenheimer-Bergman, A., Connor, S. et al. Sub-nucleosomal organization in urine cell-free DNA. bioRxiv. (2019). https://www.biorxiv.org/content/biorxiv/early/2019/07/11/696633.full.pdf.
85. 85.Burnham, P., Dadhania, D., Heyang, M., Chen, F., Westblade, L. F., Suthanthiran, M. et al. Urinary cell-free DNA is a versatile analyte for monitoring infections of the urinary tract. Nat. Commun. 9, 2412 (2018).
PubMed PubMed Central Article CAS Google Scholar
86. 86.Cook, L., Starr, K., Boonyaratanakornkit, J., Hayden, R., Sam, S. S. & Caliendo, A. M. Does size matter? Comparison of wxtraction yields for different-sized DNA fragments by seven different routine and four new circulating cell-free extraction methods. J. Clin. Microbiol. 56, e01061–18 (2018).
CAS PubMed PubMed Central Article Google Scholar
87. 87.Markus, H., Contente-Cuomo, T., Farooq, M., Liang, W. S., Borad, M. J., Sivakumar, S. et al. Evaluation of pre-analytical factors affecting plasma DNA analysis. Sci. Rep. 8, 7375 (2018).
PubMed PubMed Central Article CAS Google Scholar
88. 88.Snyder, M. W., Kircher, M., Hill, A. J., Daza, R. M. & Shendure, J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164, 57–68 (2016).
CAS PubMed PubMed Central Article Google Scholar
89. 89.Cheng, T. H. T., Lui, K. O., Peng, X. L., Cheng, S. H., Jiang, P., Chan, K. C. A. et al. DNase1 does not appear to play a major role in the fragmentation of plasma DNA in a knockout mouse model. Clin. Chem. 64, 406–408 (2018).
CAS PubMed Article PubMed Central Google Scholar
90. 90.Henikoff, S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 9, 15–26 (2008).
CAS PubMed Article PubMed Central Google Scholar
91. 91.Ivanov, M., Baranova, A., Butler, T., Spellman, P. & Mileyko, V. Non-random fragmentation patterns in circulating cell-free DNA reflect epigenetic regulation. BMC Genomics. 16, S1 (2015).
PubMed PubMed Central Article Google Scholar
92. 92.Ulz, P., Thallinger, G. G., Auer, M., Graf, R., Kashofer, K., Jahn, S. W. et al. Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat. Genet. 48, 1273–1278 (2016).
CAS PubMed Article PubMed Central Google Scholar
93. 93.Sun, K., Jiang, P., Cheng, S. H., Cheng, T. H. T., Wong, J., Wong, V. W. S. et al. Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin. Genome Res. 29, 418–427 (2019).
CAS PubMed PubMed Central Article Google Scholar
94. 94.Valouev, A., Johnson, S. M., Boyd, S. D., Smith, C. L., Fire, A. Z. & Sidow, A. Determinants of nucleosome organization in primary human cells. Nature 474, 516–520 (2011).
CAS PubMed PubMed Central Article Google Scholar
95. 95.Teif, V. B., Vainshtein, Y., Caudron-Herger, M., Mallm, J. P., Marth, C., Hofer, T. et al. Genome-wide nucleosome positioning during embryonic stem cell development. Nat. Struct. Mol. Biol. 19, 1185–1192 (2012).
CAS PubMed Article PubMed Central Google Scholar
96. 96.Ulz, P., Perakis, S., Zhou, Q., Moser, T., Belic, J., Lazzeri, I. et al. Inference of transcription factor binding from cell-free DNA enables tumor subtype prediction and early detection. Nat. Commun. 10, 4666 (2019).
CAS PubMed PubMed Central Article Google Scholar
97. 97.Jaenisch, R. & Bird, A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245–254 (2003).
CAS PubMed Article PubMed Central Google Scholar
98. 98.Yong, W. S., Hsu, F. M. & Chen, P. Y. Profiling genome-wide DNA methylation. Epigenetics Chromatin. 9, 26 (2016).
PubMed PubMed Central Article CAS Google Scholar
99. 99.Kurdyukov, S. & Bullock, M. DNA methylation analysis: choosing the right method. Biology (Basel). 5, (2016). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4810160/pdf/biology-05-00003.pdf.
100. 100.Liu, Z., Wang, Z., Jia, E., Ouyang, T., Pan, M., Lu, J. et al. Analysis of genome-wide in cell free DNA methylation: progress and prospect. Analyst. 144, 5912–5922 (2019).
CAS PubMed Article PubMed Central Google Scholar
101. 101.Shen, S. Y., Singhania, R., Fehringer, G., Chakravarthy, A., Roehrl, M. H. A., Chadwick, D. et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 563, 579–583 (2018).
CAS PubMed Article PubMed Central Google Scholar
102. 102.De Koker, A., Van Paemel, R., De Wilde, B., De Preter, K.& Callewaert, N. A versatile method for circulating cell-free DNA methylome profiling by reduced representation bisulfite sequencing. bioRxiv. (2019). https://www.biorxiv.org/content/biorxiv/early/2019/06/11/663195.full.pdf.
103. 103.Gai, W. & Sun, K. Epigenetic biomarkers in cell-free DNA and applications in liquid biopsy. Genes (Basel) 10, (2019). https://doi.org/10.3390/genes10010032.
104. 104.Warton, K., Mahon, K. L. & Samimi, G. Methylated circulating tumor DNA in blood: power in cancer prognosis and response. Endocr. Relat. Cancer 23, R157–R171 (2016).
CAS PubMed PubMed Central Article Google Scholar
105. 105.Li, L., Fu, K., Zhou, W. & Snyder, M. Applying circulating tumor DNA methylation in the diagnosis of lung cancer. Precis. Clin. Med. 2, 45–56 (2019).
Article Google Scholar
106. 106.Wu, A., Cremaschi, P., Wetterskog, D., Conteduca, V., Franceschini, G. M., Kleftogiannis, D. et al. Genome-wide plasma DNA methylation features of metastatic prostate cancer. J. Clin. Invest. 130, 1991–2000 (2020).
CAS PubMed PubMed Central Article Google Scholar
107. 107.Jones, P. A., Ohtani, H., Chakravarthy, A. & De Carvalho, D. D. Epigenetic therapy in immune-oncology. Nat. Rev. Cancer 19, 151–61. (2019).
CAS PubMed Article PubMed Central Google Scholar
108. 108.Emran, A. A., Chatterjee, A., Rodger, E. J., Tiffen, J. C., Gallagher, S. J., Eccles, M. R. et al. Targeting DNA methylation and EZH2 activity to overcome melanoma resistance to immunotherapy. Trends Immunol. 40, 328–344 (2019).
CAS PubMed Article PubMed Central Google Scholar
109. 109.Xu, R. H., Wei, W., Krawczyk, M., Wang, W., Luo, H., Flagg, K. et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 16, 1155–1161 (2017).
CAS PubMed Article PubMed Central Google Scholar
110. 110.Roadmap Epigenomics, C., Kundaje, A., Meuleman, W., Ernst, J., Bilenky, M., Yen, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Article CAS Google Scholar
111. 111.Poon, L. L., Leung, T. N., Lau, T. K., Chow, K. C. & Lo, Y. M. Differential DNA methylation between fetus and mother as a strategy for detecting fetal DNA in maternal plasma. Clin. Chem. 48, 35–41 (2002).
CAS PubMed Article PubMed Central Google Scholar
112. 112.Lun, F. M., Chiu, R. W., Sun, K., Leung, T. Y., Jiang, P., Chan, K. C. et al. Noninvasive prenatal methylomic analysis by genomewide bisulfite sequencing of maternal plasma DNA. Clin. Chem. 59, 1583–1594 (2013).
CAS PubMed Article PubMed Central Google Scholar
113. 113.Lehmann-Werman, R., Neiman, D., Zemmour, H., Moss, J., Magenheim, J., Vaknin-Dembinsky, A. et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl Acad. Sci. USA 113, E1826–E1834 (2016).
CAS PubMed Article PubMed Central Google Scholar
114. 114.Sun, K., Jiang, P., Chan, K. C., Wong, J., Cheng, Y. K., Liang, R. H. et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl Acad. Sci. USA 112, E5503–E5512 (2015).
CAS PubMed Article PubMed Central Google Scholar
115. 115.Gai, W., Ji, L., Lam, W. K. J., Sun, K., Jiang, P., Chan, A. W. H. et al. Liver- and colon-specific DNA methylation markers in plasma for investigation of colorectal cancers with or without liver metastases. Clin. Chem. 64, 1239–1249 (2018).
CAS PubMed Article PubMed Central Google Scholar
116. 116.Liao, J. B. Viruses and human cancer. Yale J. Biol. Med. 79, 115–122 (2006).
CAS PubMed PubMed Central Google Scholar
117. 117.Farrell, P. J. Epstein-Barr virus and cancer. Annu. Rev. Pathol. 14, 29–53 (2019).
CAS PubMed Article PubMed Central Google Scholar
118. 118.Borsetto, D., Cheng, J., Payne, K., Nankivell, P., Batis, N., Rao, K. et al. Surveillance of HPV-positive head and neck squamous cell carcinoma with circulating and salivary DNA biomarkers. Crit. Rev. Oncog. 23, 235–245 (2018).
PubMed Article PubMed Central Google Scholar
119. 119.Cheung, T. H., Yim, S. F., Yu, M. Y., Worley, M. J. Jr, Fiascone, S. J., Chiu, R. W. K. et al. Liquid biopsy of HPV DNA in cervical cancer. J. Clin. Virol. 114, 32–36 (2019).
CAS PubMed Article PubMed Central Google Scholar
120. 120.Cocuzza, C. E., Martinelli, M., Sina, F., Piana, A., Sotgiu, G., Dell’Anna, T. et al. Human papillomavirus DNA detection in plasma and cervical samples of women with a recent history of low grade or precancerous cervical dysplasia. PloS ONE 12, e0188592 (2017).
PubMed PubMed Central Article CAS Google Scholar
121. 121.Jensen, K. K., Gronhoj, C., Jensen, D. H. & von Buchwald, C. Circulating human papillomavirus DNA as a surveillance tool in head and neck squamous cell carcinoma: a systematic review and meta-analysis. Clin. Otolaryngol. 43, 1242–1249 (2018).
CAS PubMed Article PubMed Central Google Scholar
122. 122.Ahn, S. M., Chan, J. Y., Zhang, Z., Wang, H., Khan, Z., Bishop, J. A. et al. Saliva and plasma quantitative polymerase chain reaction-based detection and surveillance of human papillomavirus-related head and neck cancer. JAMA Otolaryngol. Head Neck Surg. 140, 846–854 (2014).
PubMed PubMed Central Article Google Scholar
123. 123.Wang, Y., Springer, S., Mulvey, C. L., Silliman, N., Schaefer, J., Sausen, M. et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci. Transl. Med. 7, 293ra104 (2015).
PubMed PubMed Central Article CAS Google Scholar
124. 124.Hanna, G. J., Lau, C. J., Mahmood, U., Supplee, J. G., Mogili, A. R., Haddad, R. I. et al. Salivary HPV DNA informs locoregional disease status in advanced HPV-associated oropharyngeal cancer. Oral. Oncol. 95, 120–126 (2019).
CAS PubMed Article PubMed Central Google Scholar
125. 125.Schmidt, H., Kulasinghe, A., Allcock, R. J. N., Tan, L. Y., Mokany, E., Kenny, L. et al. A pilot study to non-invasively track PIK3CA mutation in head and neck cancer. Diagnostics (Basel). 8, (2018). https://doi.org/10.3390/diagnostics8040079.
126. 126.Lam, W. K. J., Chan, K. C. A. & Lo, Y. M. D. Plasma Epstein-Barr virus DNA as an archetypal circulating tumour DNA marker. J. Pathol. 247, 641–649 (2019).
PubMed PubMed Central Article Google Scholar
127. 127.Lo, Y. M., Chan, A. T., Chan, L. Y., Leung, S. F., Lam, C. W., Huang, D. P. et al. Molecular prognostication of nasopharyngeal carcinoma by quantitative analysis of circulating Epstein-Barr virus DNA. Cancer Res. 60, 6878–6881 (2000).
CAS PubMed PubMed Central Google Scholar
128. 128.Chen, Q. Y., Guo, S. Y., Tang, L. Q., Lu, T. Y., Chen, B. L., Zhong, Q. Y. et al. Combination of tumor volume and Epstein-Barr vrus DNA improved prognostic stratification of stage II nasopharyngeal carcinoma in the intensity modulated radiotherapy era: a large-scale cohort study. Cancer Res. Treat. 50, 861–871 (2018).
CAS PubMed Article PubMed Central Google Scholar
129. 129.Leung, S. F., Chan, K. C., Ma, B. B., Hui, E. P., Mo, F., Chow, K. C. et al. Plasma Epstein-Barr viral DNA load at midpoint of radiotherapy course predicts outcome in advanced-stage nasopharyngeal carcinoma. Ann. Oncol. 25, 1204–1208 (2014).
CAS PubMed Article PubMed Central Google Scholar
130. 130.Chen, Q., Hu, W., Xiong, H., Ying, S., Ruan, Y., Wu, B. et al. Changes in plasma EBV-DNA and immune status in patients with nasopharyngeal carcinoma after treatment with intensity-modulated radiotherapy. Diagn. Pathol. 14, 23 (2019).
PubMed PubMed Central Article Google Scholar
131. 131.Chan, K. C. A., Woo, J. K. S., King, A., Zee, B. C. Y., Lam, W. K. J., Chan, S. L. et al. Analysis of plasma Epstein-Barr virus DNA to screen for nasopharyngeal cancer. N. Engl. J. Med. 377, 513–522 (2017).
CAS PubMed Article PubMed Central Google Scholar
132. 132.He, S. S., Wang, Y., Bao, Y., Cai, X. Y., Yang, X. L., Chen, D. M. et al. Dynamic changes in plasma Epstein-Barr virus DNA load during treatment have prognostic value in nasopharyngeal carcinoma: a retrospective study. Cancer Med. 7, 1110–1117 (2018).
CAS PubMed PubMed Central Article Google Scholar
133. 133.Lv, J., Chen, Y., Zhou, G., Qi, Z., Tan, K. R. L., Wang, H. et al. Liquid biopsy tracking during sequential chemo-radiotherapy identifies distinct prognostic phenotypes in nasopharyngeal carcinoma. Nat. Commun. 10, 3941 (2019).
PubMed PubMed Central Article CAS Google Scholar
134. 134.Lam, W. K. J., Jiang, P., Chan, K. C. A., Cheng, S. H., Zhang, H., Peng, W. et al. Sequencing-based counting and size profiling of plasma Epstein-Barr virus DNA enhance population screening of nasopharyngeal carcinoma. Proc. Natl Acad. Sci. USA 115, E5115–E5124 (2018).
CAS PubMed Article PubMed Central Google Scholar
135. 135.Qiu, M. Z., He, C. Y., Lu, S. X., Guan, W. L., Wang, F., Wang, X. J. et al. Prospective observation: clinical utility of plasma Epstein-Barr virus DNA load in EBV-associated gastric carcinoma patients. Int. J. Cancer. 146, 272–280 (2019).
PubMed Article CAS PubMed Central Google Scholar
136. 136.To, E. W., Chan, K. C., Leung, S. F., Chan, L. Y., To, K. F., Chan, A. T. et al. Rapid clearance of plasma Epstein-Barr virus DNA after surgical treatment of nasopharyngeal carcinoma. Clin. Cancer Res. 9, 3254–3259 (2003).
CAS PubMed PubMed Central Google Scholar
137. 137.Lampignano, R., Neumann, M. H. D., Weber, S., Kloten, V., Herdean, A., Voss, T. et al. Multicenter evaluation of circulating cell-free DNA extraction and downstream analyses for the development of standardized (Pre)analytical work flows. Clin. Chem. 66, 149–160 (2019).
Article Google Scholar
138. 138.Meddeb, R., Dache, Z. A. A., Thezenas, S., Otandault, A., Tanos, R., Pastor, B. et al. Quantifying circulating cell-free DNA in humans. Sci. Rep. 9, 5220 (2019).
PubMed PubMed Central Article CAS Google Scholar
139. 139.Anfossi, S., Babayan, A., Pantel, K. & Calin, G. A. Clinical utility of circulating non-coding RNAs—an update. Nat. Rev. Clin. Oncol. 15, 541–563 (2018).
PubMed Article PubMed Central Google Scholar
140. 140.Cohen, J. D., Li, L., Wang, Y., Thoburn, C., Afsari, B., Danilova, L. et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018).
CAS PubMed PubMed Central Article Google Scholar
141. 141.Pantel, K. & Alix-Panabieres, C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol. Med. 16, 398–406 (2010).
142. 142.Murtaza, M. & Caldas, C. Nucleosome mapping in plasma DNA predicts cancer gene expression. Nat. Genet. 48, 1105–1106 (2016).
143. 143.Forshew, T., Murtaza, M., Parkinson, C., Gale, D., Tsui, D. W. Y., Kaper, F. et al. Noninvasive Identification and Monitoring of Cancer Mutations by Targeted Deep Sequencing of Plasma DNA. Sci. Trans. Med 4, 136ra68 (2012).
144. 144.De Mattos-Arruda, L., Weigelt, B., Cortes, J., Won, H. H., Ng, C. K. Y., Nuciforo, P. et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann. Oncol. 25, 1729–1735 (2014).
145. 145.Khan, K. H., Cunningham, D., Werner, B., Vlachogiannis, G., Spiteri, I., Heide, T. et al. Longitudinal Liquid Biopsy and Mathematical Modeling of Clonal Evolution Forecast Time to Treatment Failure in the PROSPECT-C Phase II Colorectal Cancer Clinical Trial. Cancer Discov. 8, 1270–1285 (2018).
146. 146.Jamal-Hanjani, M., Wilson, G. A., Horswell, S., Mitter, R., Sakarya, O., Constantin, T. et al. Detection of ubiquitous and heterogeneous mutations in cell-free DNA from patients with early-stage non-small-cell lung cancer. Ann. Oncol. 27, 862–867 (2016).
147. 147.Gao, J., Wang, H., Zang, W., Li, B., Rao, G., Li, L. et al. Circulating tumor DNA functions as an alternative for tissue to overcome tumor heterogeneity in advanced gastric cancer. Cancer Sci. 108, 1881–1887 (2017).
148. 148.Bernard-Tessier, A., Jeannot, E., Guenat, D., Debernardi, A., Michel, M., Proudhon, C. et al. Clinical Validity of HPV Circulating Tumor DNA in Advanced Anal Carcinoma: An Ancillary Study to the Epitopes-HPV02 Trial. Clin. Cancer Res. 25, 2109–2115 (2019).
149. 149.Cabel, L., Jeannot, E., Bieche, I., Vacher, S., Callens, C., Bazire, L. et al. Prognostic Impact of Residual HPV ctDNA Detection after Chemoradiotherapy for Anal Squamous Cell Carcinoma. Clin. Cancer Res. 24, 5767–5771 (2018).
150. 150.Carow, K., Golitz, M., Wolf, M., Hafner, N., Jansen, L., Hoyer, H. et al. Viral-Cellular DNA Junctions as Molecular Markers for Assessing Intra-Tumor Heterogeneity in Cervical Cancer and for the Detection of Circulating Tumor DNA. Int. J. Mol. Sci. 18, 2032 (2017).
151. 151.Kang, Z., Stevanovic, S., Hinrichs, C. S. & Cao, L. Circulating Cell-free DNA for Metastatic Cervical Cancer Detection, Genotyping, and Monitoring. Clin. Cancer Res. 23, 6856–6862 (2017)
152. 152.Jeannot, E., Becette, V., Campitelli, M., Calmejane, M. A., Lappartient, E., Ruff, E. et al. Circulating human papillomavirus DNA detected using droplet digital PCR in the serum of patients diagnosed with early stage human papillomavirus-associated invasive carcinoma. J Pathol. Clin. Res. 2, 201–209 (2016).
153. 153.Sathish, N., Abraham, P., Peedicayil, A., Sridharan, G., John, S., Shaji, R. V. et al. HPV DNA in plasma of patients with cervical carcinoma. J. Clin. Virol. 31, 204–209 (2004).
154. 154.Campitelli, M., Jeannot, E., Peter, M., Lappartient, E., Saada, S., de la Rochefordiere, A. et al. Human papillomavirus mutational insertion: specific marker of circulating tumor DNA in cervical cancer patients. PloS ONE. 7, e43393 (2012).
155. 155.Mazurek, A. M., Rutkowski, T., Fiszer-Kierzkowska, A., Malusecka, E. & Skladowski, K. Assessment of the total cfDNA and HPV16/18 detection in plasma samples of head and neck squamous cell carcinoma patients. Oral oncology 54, 36–41 (2016).
156. 156.Capone, R. B., Pai, S. I., Koch, W. M., Gillison, M. L., Danish, H. N., Westra, W. H. et al. Detection and quantitation of human papillomavirus (HPV) DNA in the sera of patients with HPV-associated head and neck squamous cell carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research 6, 4171–4175 (2000).
157. 157.Dahlstrom, K. R., Li, G., Hussey, C. S., Vo, J. T., Wei, Q., Zhao, C. et al. Circulating human papillomavirus DNA as a marker for disease extent and recurrence among patients with oropharyngeal cancer. Cancer 121, 3455–3464 (2015).
158. 158.Cao, H., Banh, A., Kwok, S., Shi, X., Wu, S., Krakow, T. et al. Quantitation of human papillomavirus DNA in plasma of oropharyngeal carcinoma patients. International journal of radiation oncology, biology, physics 82, e351–e358 (2012).
转载自:
Keller, L., Belloum, Y., Wikman, H. et al.
Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond.
Br J Cancer 124, 345–358 (2021).
https://rdcu.be/celxF




发表评论 取消回复